Pathogens exert one of the strongest selective pressures on plant populations. This leads to progressive changes in the genetic makeup of both the pathogen as well the plant, which is usually referred to as an arms race. Fungal pathogens are one of the highly evolved groups of microorganisms that affect various plant species and strongly differ in important life history traits such as dispersal mechanisms, type of reproduction and modes of parasitism. Economic damage due to fungal diseases in crops is estimated to be over 200 billion USD.
The Brassicaceae family consists of over 40 crop species that are grown worldwide as oilseeds, vegetables, or condiments. Fungal and oomycete pathogens such as Alternaria brassicae, Sclerotinia sclerotiorum, and Albugo candida are some of the major destabilizers of yield in the mustard crops. Our lab focusses on one of the major fungal diseases of the Brassicaceae family, Alternaria Blight or Alternaria Leaf spot caused by Alternaria brassicae. No source of resistance is known in any of the cultivated species of Brassicas. We aim to understand both the mechanisms of pathogenesis and disease resistance and use the knowledge gained therein to assist disease resistance breeding programmes. The research projects ongoing in the lab are listed below.
Host Resistance - 1. Deciphering the genetic basis of the disease response framework of Brassica juncea to Alternaria brassicae
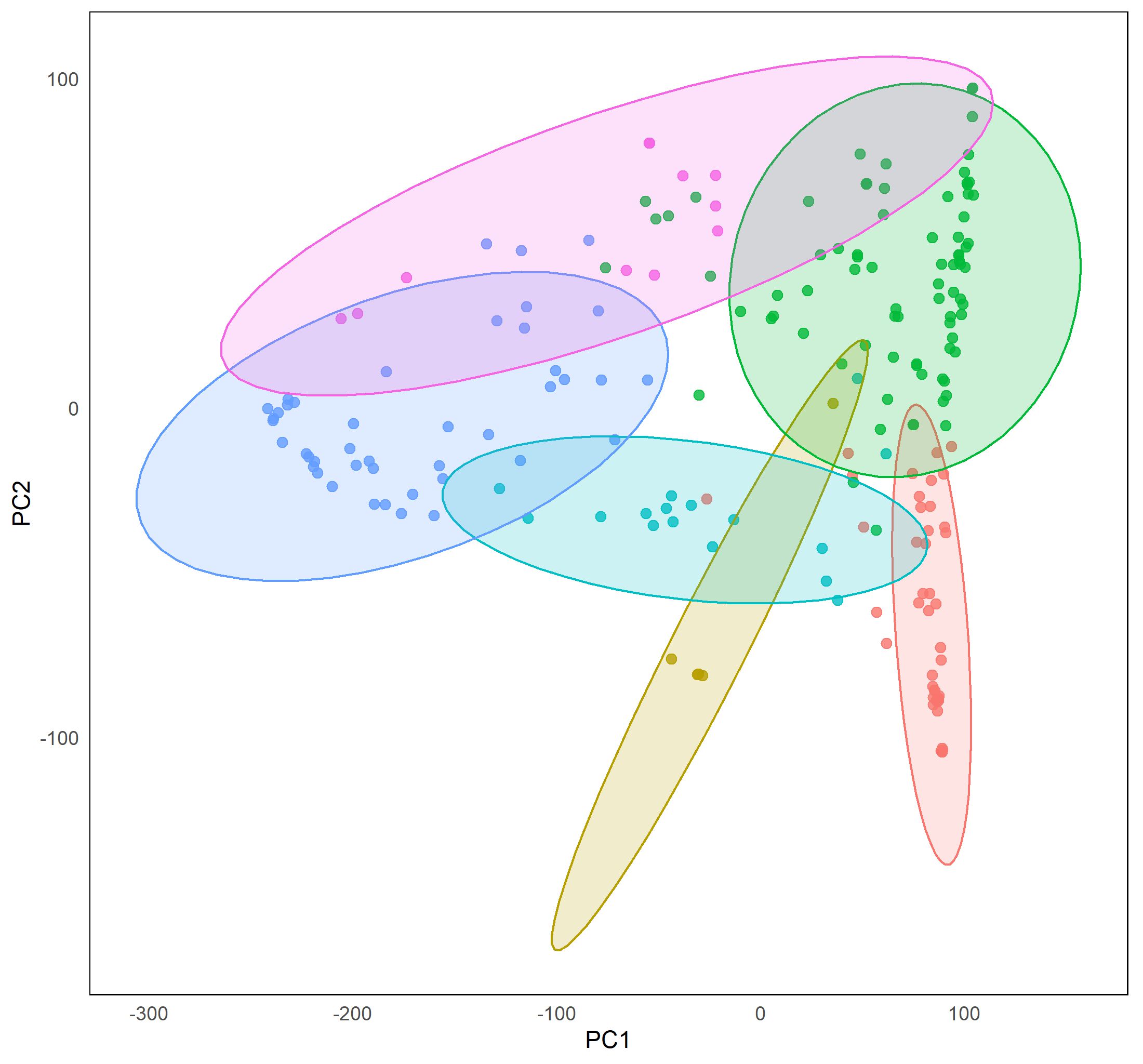
Our previous research on the genetics of resistance to Alternaria in Arabidopsis revealed that the resistance is quantitative and multigenic (Rajarammohan et al., 2017; 2018). Quantitative disease resistance may be conditioned by genes that regulate morphological and developmental phenotypes, are involved in basal defense, synthesize defense-related secondary metabolites, are involved in defense signal transduction, and are direct receptors of molecules secreted by the pathogen. Therefore, we aim to stduy endophenotypes or cellular phenotypes of disease resistance so as to build a disease response framework of B. juncea. In this direction we are trying to unravel the genetic basis of endophenotypes such as secreted peroxidase (sPOX) activity, ROS burst, MAP kinase signaling, lignin accumulation, ethylene production etc. We are using a panel of 302 B. juncea accessions collected worldwide for this purpose. This project is funded by the DST-INSPIRE faculty scheme, Department of Science & Technology, Government of India.
Host Resistance - 2. Secondary/tertiary gene pools as a source of resistance to A. brassicae
The related but wild relatives of many crop species are known to be rich reservoir of disease resistance genes. We have chosen two related species of B. juncea - one cultivated (Brassica carinata) and one wild (Arabidopsis thaliana) species to prospect for resistance genes/QTLs. B. carinata or Ethiopian mustard is an underutilized crop that is majorly grown in North-eastern Africa for its leaf (as vegetable) and seeds (as condiments). However, it is now being projected as a biofuel crop in parts of the US and Europe. We believe that the environmental resilience and disease resistance of B. carinata as compared to the other cultivated Brassicas of the U’s triangle is particularly interesting. As a start, in collaboration with scientists at CGMCP, University of Delhi South Campus, we have sequenced HC20, an accession of B. carinata that we found to be resistant against multiple pathogens of the Brassicaceae. Further, we plan to identify and mobilize the disease resistance loci from B. carinata to B. juncea via interspecific hybridization. This work is a part of the DBT-UDSC partnership project funded by Department of Biotechnology, Government of India.
The global collections of Arabidopsis lack representation from the Indian subcontinent. A group of scientists at NBRI, Lucknow have been involved in collecting and cataloging Arabidopsis accessions from the Western Himalayan region in India. We are actively collaborating with them to sequence the accessions, and to identify novel disease resistance genes from this resource. This project is funded by Department of Biotechnology, Government of India.
Pathogenesis - 1. Molecular mechanisms underlying pathogenesis of A. brassicae
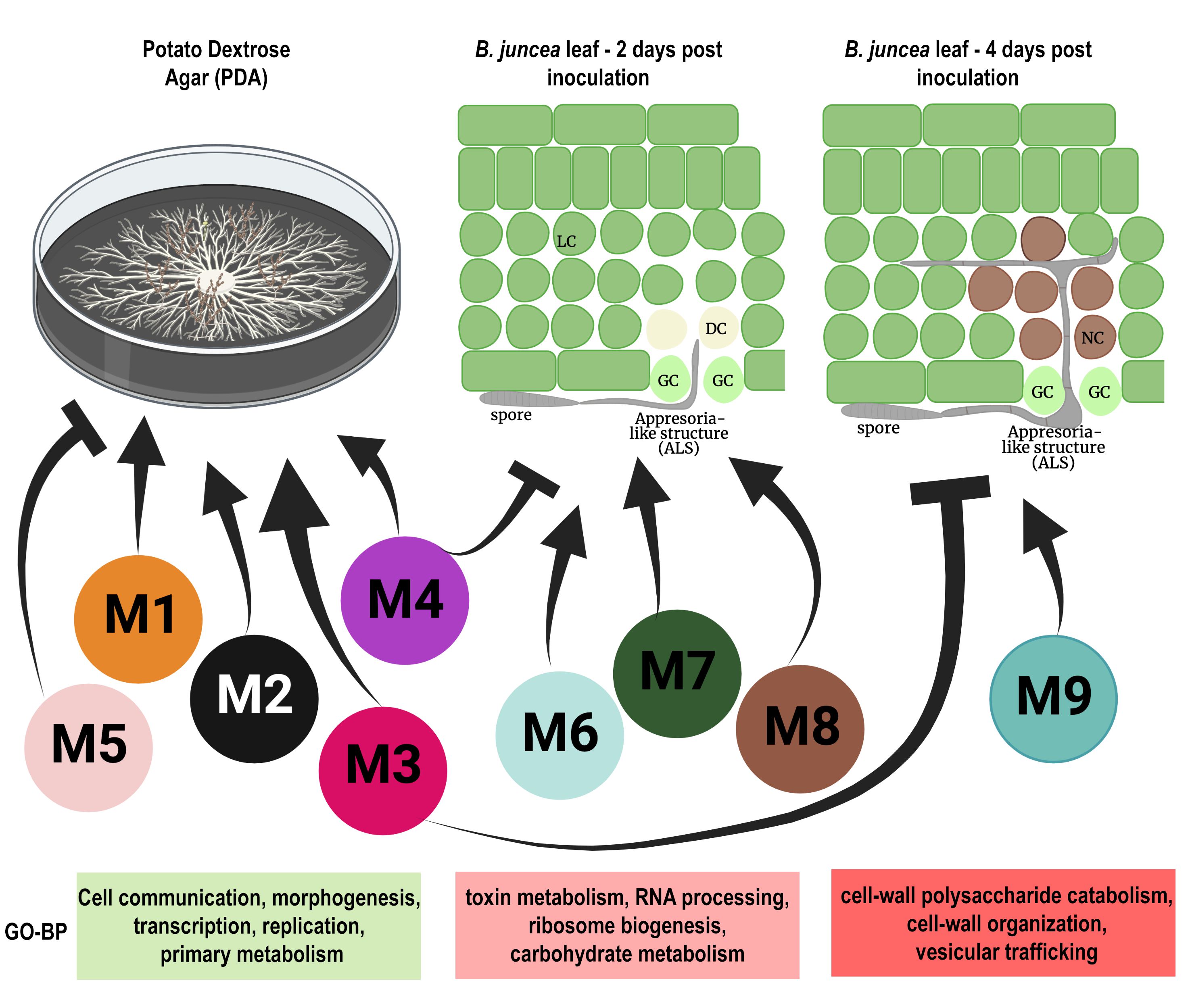
Alternaria brassicae is a necrotrophic pathogen that is able to infect almost all members of the Brassicaceae family. A. brassicae is one of the dominant invasive species on oleiferous Brassicas. Understanding the molecular mechanisms employed by the pathogen during the infection process is essential for identifying novel targets for disease management. However, very little is known about the mechanisms/pathways underlying the pathogenesis of A. brassicae. We generated a whole-transcriptome dataset of A. brassicae during growth (on potato dextrose agar plates) and infection (on B. juncea var. Varuna) and used it to derive some basic information about the pathogenesis of A. brassicae. Future studies would involve studying the expression profile of A. brassicae at higher temporal resolution in different host species. This project is funded by a Core Research Grant (CRG) from Science and Engineering Research Board (SERB), Government of India.
Pathogenesis - 2. Identifying effectors of A. brassicae and dissecting their role in pathogenesis
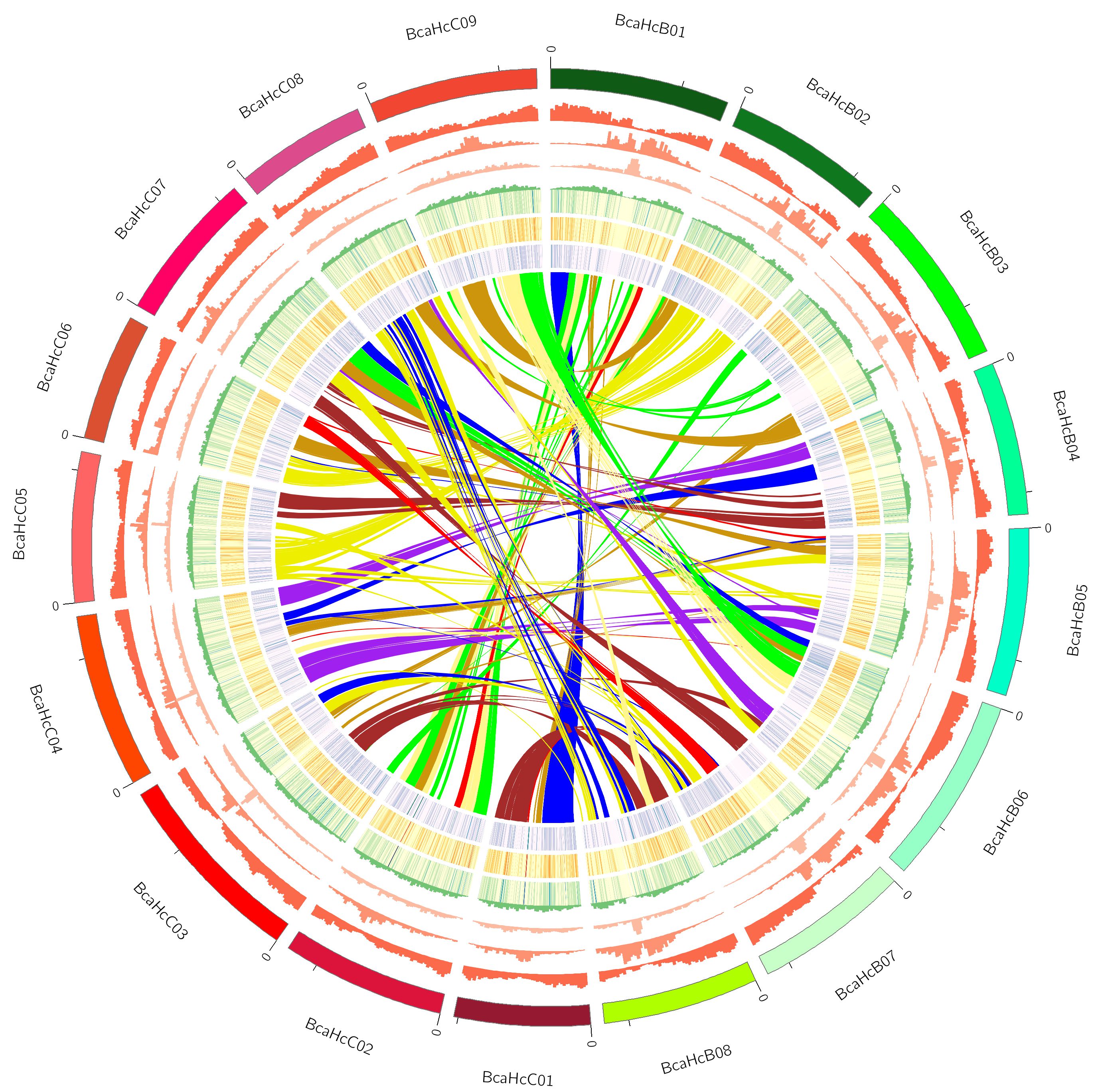
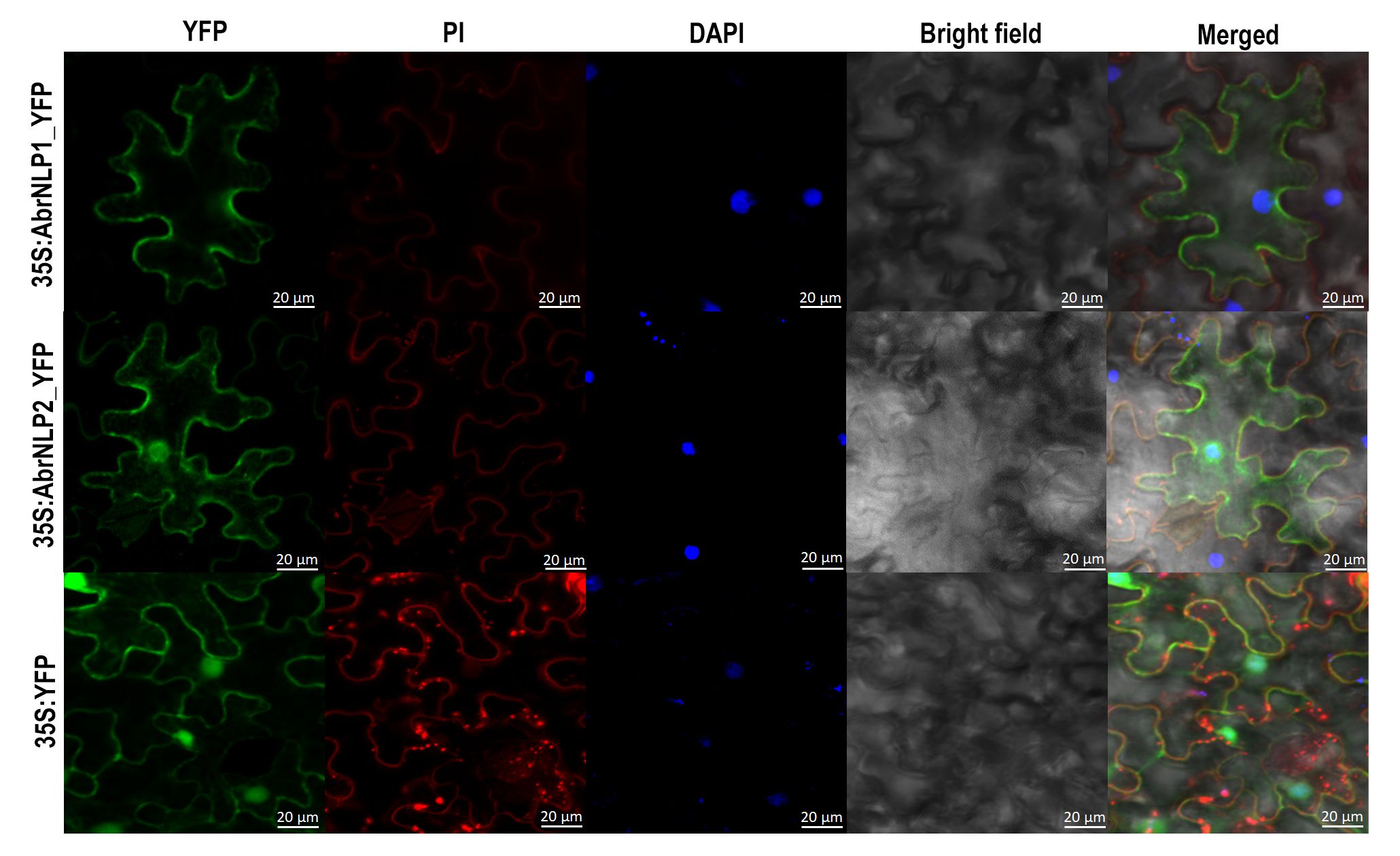
We have sequenced and produced a highly-contiguous genome assembly of A. brassicae (Rajarammohan et al., 2019). Using various gene prediction and annotation pipelines, we have been able to predict and annotate various CAZy (Carbohydrate active enzymes), effectors, and secondary metabolite clusters (Rajarammohan et al., 2020). Furthermore, we also aim to characterise the identified effectors for their role in pathogenesis. As a start, we characterised the NLPs (necrosis and ethylene-inducing peptide-like proteins) from A. brassicae. We found that AbrNLPs are functionally and spatially (subcellular location) distinct and may play different but important roles during the pathogenesis of A. brassicae. This project is funded by a Core Research Grant (CRG) from Science and Engineering Research Board (SERB), Government of India.
Pathogenesis - 3. Exploring the genetic diversity of A. brassicae across India to gain insights into the evolutionary history of the pathogen
Understanding a pathogen’s genetic diversity is crucial to the effectiveness and durability of host resistance. Population genetics of plant pathogens has enormous potential to unravel the evolutionary forces acting on pathogen population, and this information can be used to develop strategies of resistance breeding. We aim to catalogue the genetic diversity of various isolates of A. brassicae across India at the genomic level and analyse their implications in pathogenesis.